Research of Our Team
I. Plant innate immunity
Plant evolved the innate immune system to combat against microbial pathogens. We investigate the mechanism by which plant immunity combats plant pathogens with a focus on plant defense hormones such as salicylic acid, jasmonate, and ethylene and MAP kinases. More recently, we investigate transcriptomes and proteomes of both the plant Arabidopsis thaliana and the bacterial pathogen Pseudomonas syringae during the interaction, which reveals how plant immunity affects bacterial metabolism such as translation and iron acquisition and bacterial virulence thereby controlling pathogen growth. This generates a number of hypotheses to be tested in the future.
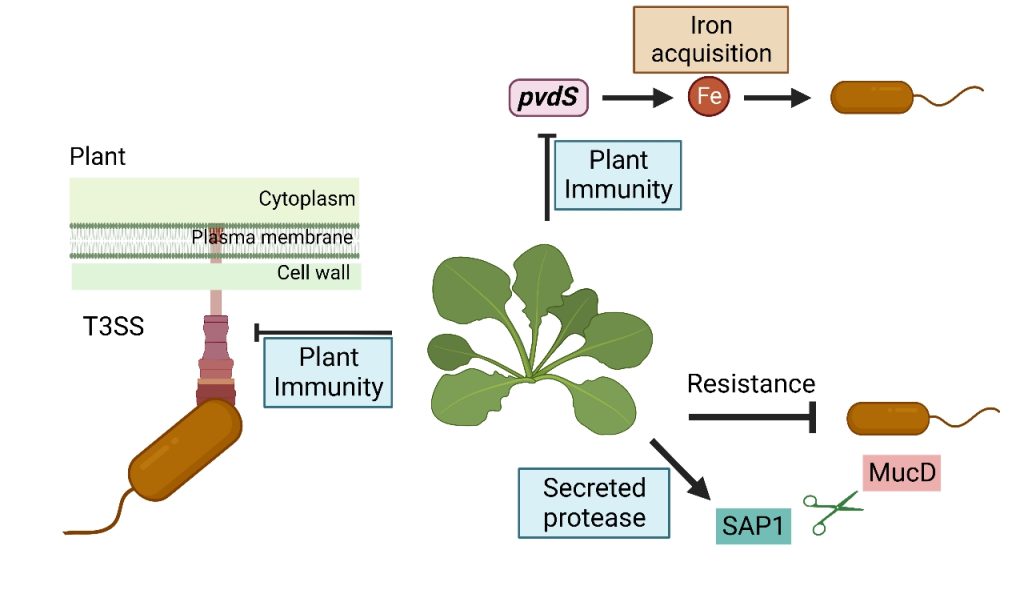
Plants can inhibit the growth of invading bacteria, but the mechanism remains obscure. Further, such defense often comes at the expense of plant growth. We discovered that an evolutionarily conserved plant protein serves as a ‘molecular scissors’ that cuts a highly conserved bacterial protein important for virulence and thereby directly suppressing bacterial growth. Artificially boosting expression of the molecular scissors increased plant resistance but did not trigger immune activation associated with plant growth retardation. Thus, our finding suggests an approach for increasing pathogen resistance without compromising plant yields.
In response to local pathogen infection, plants are capable of increasing disease resistance in distant leaves, a phenomenon termed systemic acquired resistance (SAR). We investigate the molecular mechanism of SAR. We are interested in how plants develop SAR in natural conditions where a diverse microbes associate with plants and microbe-produced metabolites (including SAR-related chemicals) would influence the state of SAR.
Trade-offs between stress responses to physical and biological factors are thought to contribute to prioritizing responses to one stress over the other thereby increasing plant fitness in response to individual stresses. However, this does not explain if and how this crosstalk is beneficial under conditions where a plant would encounter both types of stress simultaneously, a situation which is frequent in nature. We investigate how plants cope with multiple stress environments.
Related publications (since 2017)
- Entila F, Han X, Mine A, Schulze-Lefert P, Tsuda K*. Commensal lifestyle regulated by a negative feedback loop between Arabidopsis ROS and the bacterial T2SS. Nature Communications, 15:456 (2024) . 中文介绍
- Hou S, Tsuda K*: Salicylic acid and jasmonic acid crosstalk in plant immunity. Essays in Biochemistry, 66: 647-656 (2022)
- Nakano M, Omae N, Tsuda K*: Inter-organismal phytohormone networks in plant-microbe interactions. Current Opinion in Plant Biology, 68: 102258 (2022)
- Winkelmüller TM, Entila F, Anver S, Piasecka A, Song B, Dahms E, Sakakibara H, Gan X, Kułak K, Sawikowska A, Krajewski P, Tsiantis M, Garrido-Oter R, Fukushima K, Schulze-Lefert P, Laurent S, Bednarek P, Tsuda K*: Gene expression evolution in pattern-triggered immunity within Arabidopsis thaliana and across Brassicaceae species. Plant Cell, 33: 1863-1887 (2021)
- Nobori T, Wang Y, Wu J, Christina Stolze S, Tsuda T, Finkemeier I, Nakagami H, Tsuda K*: Multidimensional gene regulatory landscape of a bacterial pathogen in plants. Nature Plants, 6: 883-896 (2020)
- Wang Y, Garrido-Oter R, Wu J, Winkelmuller TM, Agler M, Colby T, Nobori T, Kemen E, Tsuda K*: Site-specific cleavage of bacterial MucD by secreted proteases mediates antibacterial resistance in Arabidopsis. Nature Communications, 10: 2853 (2019)
- Berens ML, Wolinska KW, Spaepen S, Ziegler J, Nobori T, Nair A, Krüler V, Winkelmüller TM, Wang Y, Mine A, Becker D, Garido-Oter R, Schulze-Lefert P*, Tsuda K*: Balancing trade-offs between biotic and abiotic stress responses through leaf age-dependent variation in stress hormone crosstalk. Proceedings of the National Academy of Sciences USA, 116: 2364-2373 (2019)
- Wang Y, Schuck S, Wu J, Yang P, Döring AC, Zeier J*, Tsuda K*: A MPK3/6-WRKY33-ALD1-Pipecolic acid Regulatory Loop Contributes to Systemic Acquired Resistance. Plant Cell, 10: 2480-2494 (2018)
- Mine A, Seyfferth C, Kracher B, Berens ML, Becker D, Tsuda K*: The Defense Phytohormone Signaling Network Enables Rapid, High-amplitude Transcriptional Reprogramming During Effector-Triggered Immunity. Plant Cell, 30: 1199-1219 (2018)
- Nobori T, Mine A, Tsuda K*: Molecular networks in plant-pathogen holobiont. FEBS Letters, 592: 1937-1953 (2018)
- Nobori T, Velásquez AC, Wu J, Kvitko BH, Kremer JM, Wang Y, He SY*, Tsuda K*: Transcriptome landscape of a bacterial pathogen under plant immunity. Proceedings of the National Academy of Sciences USA,115: E3055-E3064 (2018)
- Mine A, Nobori T†, Salazar-Rondon MC†, Winkelmüller TM, Anver S, Becker D, Tsuda K*: An incoherent feed-forward loop mediates robustness and tunability in a plant immune network. EMBO Reports, 18: 464-476 (2017)
II. Plant microbiota
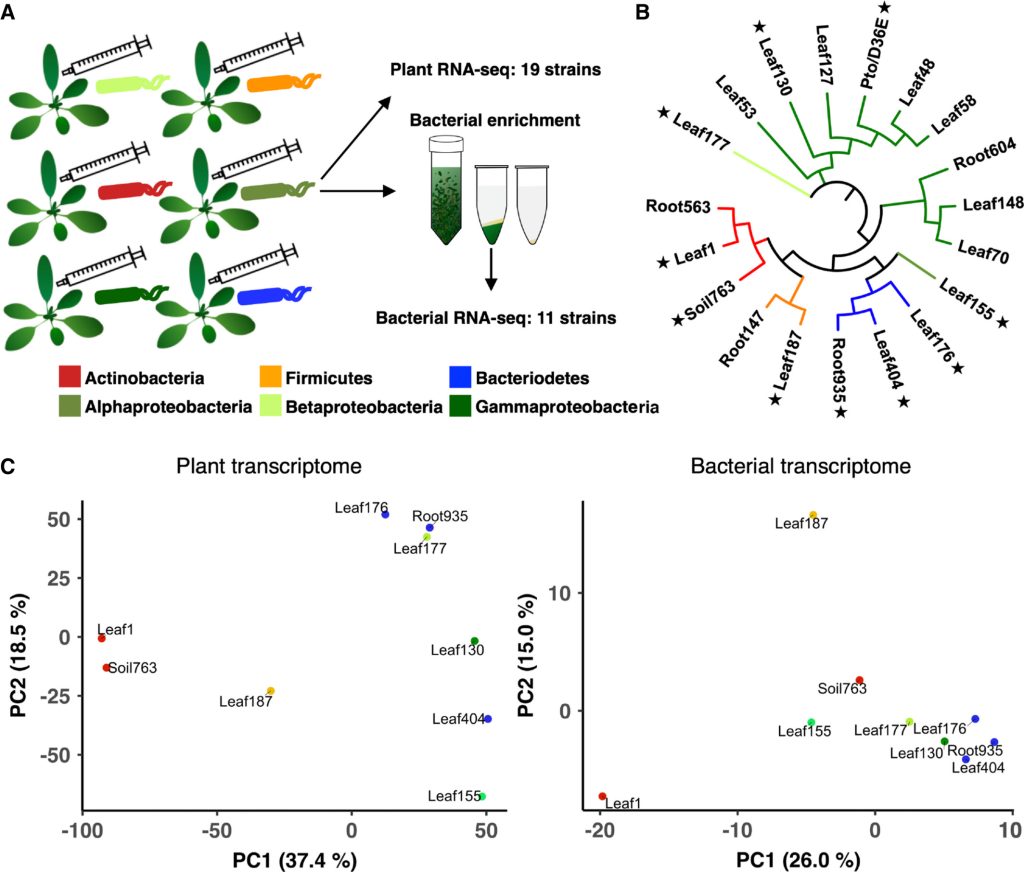
Plant evolved the innate immune system to control not only microbial pathogens but also the structure and function of plant microbiota which promote plant health. We investigate the interaction between plants and plant microbiota. Key questions include: how do plants control the function of plant microbiota? how do plants distinguish different microbiota members? what are plant and bacterial genetic determinants for compatible interactions which would lead to beneficial interactions for plants? We use synthetic bacterial communities isolated from A. thaliana and those from Z. mays to answer the above questions. In particular, we exploit the rich genetic resources of Z. mays available at this university.
Related publications (since 2017)
- Entila F, Han X, Mine A, Schulze-Lefert P, Tsuda K*. Commensal lifestyle regulated by a negative feedback loop between Arabidopsis ROS and the bacterial T2SS. Nature Communications, 15:456 (2024) . 中文介绍
- Nobori T, Cao Yu, Entila F, Dahms E, Tsuda Y, Garrido-Oter R, Tsuda K*: Dissecting the co-transcriptome landscape of plants and their microbiota. EMBO Reports, 23: e55380 (2022)
- Omae N, Tsuda K*: Plant-Microbiota Interactions in Abiotic Stress Environments. Molecular Plant-Microbe Interactions, 35: 511-526 (2022)
- Nakano M, Omae N, Tsuda K*: Inter-organismal phytohormone networks in plant-microbe interactions. Current Opinion in Plant Biology, 68: 102258 (2022)
- Berens ML, Wolinska KW, Spaepen S, Ziegler J, Nobori T, Nair A, Krüler V, Winkelmüller TM, Wang Y, Mine A, Becker D, Garido-Oter R, Schulze-Lefert P*, Tsuda K*: Balancing trade-offs between biotic and abiotic stress responses through leaf age-dependent variation in stress hormone crosstalk. Proceedings of the National Academy of Sciences USA, 116: 2364-2373 (2019)
- Nobori T, Tsuda K*: The plant immune system in heterogeneous environments. Current Opinion in Plant Biology, 50: 58-66 (2019)
III. Microbial Pathogenesis
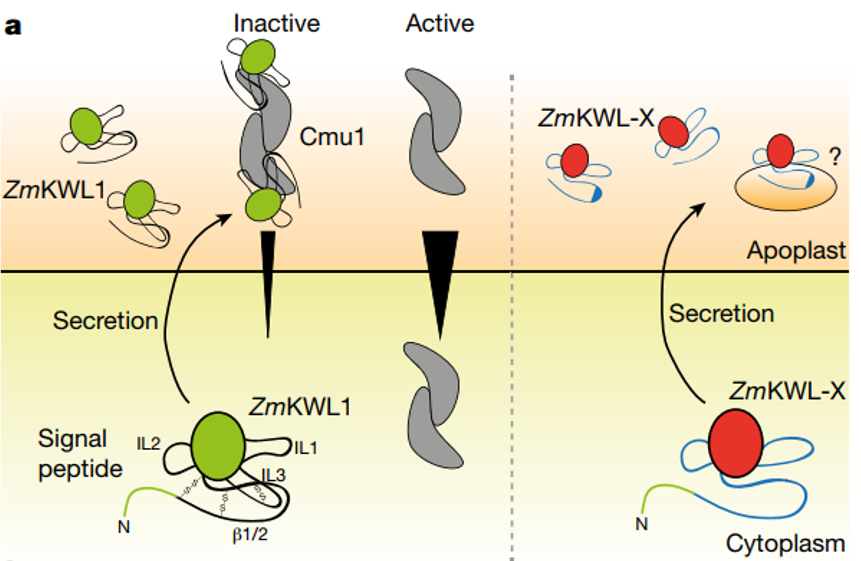
Successful plant pathogens suppress or evade plant immune responses through the secretion of virulence proteins called effectors, which either reside in the apoplastic area or translocate into plant cells to target diverse subcellular compartments. Effectors function through a variety of mechanisms: they can modify the pathogen surfaces, inactivate plant defense molecules and reprogram host physiology or metabolism to promote pathogenesis. In addition, some plant pathogens can also interfere with plant hormone signaling pathways by producing hormone mimics to inhibit plant defense response. We aim to understand the virulence strategies of the fungal pathogen Ustilago maydis and the bacterial pathogen Pseudomonas syringae pv. tomato by characterizing their secreted effector proteins.
Related publications (since 2017)
- Han X, Altegoer F, Steinchen W, Schuhmacher J, Glatter T, Giammarinaro P, Djamei A, Rensing S, Reissmann S, Kahmann R* and Bange G*: A kiwellin disarms the metabolic activity of a secreted fungal virulence factor. Nature, 565: 650-653 (2019)
- Han X and Kahmann R*: Manipulation of phytohormone pathways by effectors of filamentous plant pathogens. Front Plant Sci, 10: 822 (2019)
- Mine A, Berens ML, Nobori T, Anver S, Fukumoto K, Winkelmüller TM, Takeda A, Becker D, Tsuda K*: Pathogen exploitation of an abscisic acid- and jasmonate-inducible MAPK phosphatase and its interception by Arabidopsis immunity. Proceedings of the National Academy of Sciences USA, 114: 7456-7461 (2017)
IV. Evolution of the plant immune system
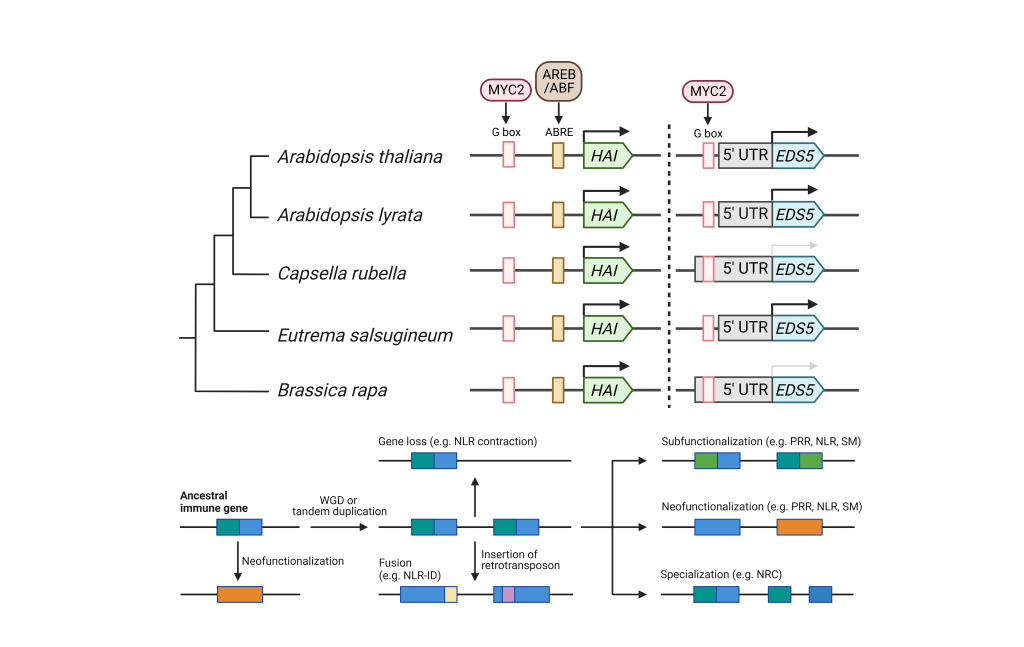
The evolutionary biologist Theodosius Dobzhansky says that “Nothing in biology makes sense except in the light of evolution”. The plant innate immune system co-evolved with environmental microbes. We investigate how plants evolve in terms of plant immunity with a particular focus on transcriptome evolution as well as the evolution of plant defense hormone biosynthesis and signaling.
Related publications (since 2017)
- Berens ML, Berry HM, Mine A, Argueso CT, Tsuda K*: Evolution of Hormone Signaling Networks in Plant Defense. Annual Review of Phytopathology, 55: 401-425 (2017)
- Han X, Tsuda K*: Evolutionary footprint of plant immunity. Current Opinion in Plant Biology, 67: 102209 (2022)
- Winkelmüller TM, Entila F, Anver S, Piasecka A, Song B, Dahms E, Sakakibara H, Gan X, Kułak K, Sawikowska A, Krajewski P, Tsiantis M, Garrido-Oter R, Fukushima K, Schulze-Lefert P, Laurent S, Bednarek P, Tsuda K*: Gene expression evolution in pattern-triggered immunity within Arabidopsis thaliana and across Brassicaceae species. Plant Cell, 33: 1863-1887 (2021)
